A Two Bit Decoder
code clojure
Saturday, July 6, 2013
The entire human genome is available as a single .2bit file here (click on “Full Data Set”, then download hg19.2bit). Unlike the stellar signal in His Master’s Voice, the 2bit format is reasonably clearly documented.
We want to write Clojure code to:
- Provide base pairs in symbolic (rather than raw binary) form as lazy sequences – i.e., sequences which need not all fit in memory at once, but can be consumed and processed as needed;
- Provide “random access” to this data selectively, e.g. by chromosome, rather than always reading through the entire file;
- Provide access to metadata encoded in the file.
The functionality to do this is posted in the jenome project on GitHub. In this post, we’ll explore this code a little; in following posts, we’ll do some investigating of the actual genome per se.
Capability (2) is provided, in part, by implementing random-access
reads from file fname of len bytes starting at offset:
(ns jenome.rafile
(:import (java.io RandomAccessFile)))
(defn read-with-offset [fname offset len]
(let [raf (RandomAccessFile. fname "r")
bb (byte-array len)]
(.seek raf offset)
(.readFully raf bb)
(.close raf)
bb))
Armed with this, we can get the .2bit file header:
(defn file-header [fname]
(let [[sig ver seqcnt resvd] (->> (read-with-offset fname 0 16)
(partition 4)
(map bytes-to-number))]
(assert (= sig 0x1A412743))
(assert (= ver 0))
(assert (= resvd 0))
seqcnt))
file-header basically just gives us the number of sequences (usually,
chromosomes) in the file, doing some sanity checks along the
way. bytes-to-number converts an arbitrary sequence of bytes to the
appropriate unsigned integer. (For brevity’s sake, I won’t show every
utility function in this blog post; the source code on GitHub is
reasonably short.)
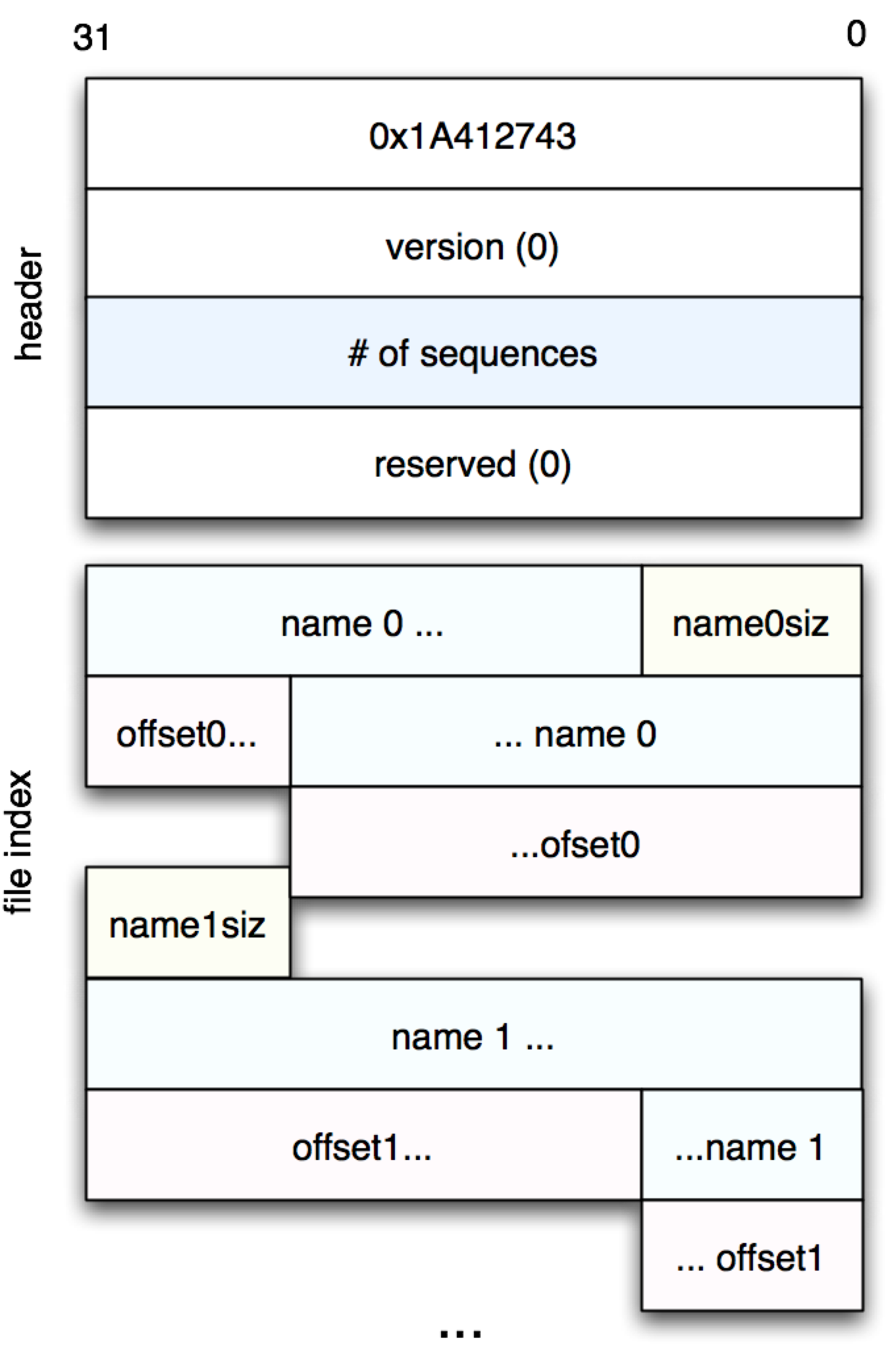
The next part of the file, as shown in Figure 1, is called the “file index,” and contains a list of sequences contained the rest of the file. It can be read as follows:
(defn file-index [fname seqcnt]
(loop [i 0
ofs 16
ret []]
(if (< i seqcnt)
(let [[nlen] (read-with-offset fname ofs 1)
name (apply str (map char (read-with-offset fname
(+ ofs 1) nlen)))
seq-offset (get32 fname (+ ofs 1 nlen))]
(recur (inc i) (+ ofs nlen 5) (conj ret [nlen name seq-offset])))
ret)))
This somewhat imperative code walks through the seqcnt sequence
portions of the index, pulling out sequence names and lengths as we
go.
It’s here that we introduce a new friend, the yeast Saccharomyces
cerevisiae (SacCer3), used since antiquity for making bread and
fermented beverages. Relatively small in comparison with the human
genome, SacCer3 will be our “unit test” organism. Available here and
checked into resources, the file can be accessed as
(def yeast (as-file (resource "sacCer3.2bit")))
(I have imported resource and as-file from clojure.java.io; again, see
the source code.)
Our index-reading code yields:
(let [seqcnt (file-header yeast)]
(file-index yeast seqcnt))
;=>
[[4 "chrI" 191]
[5 "chrII" 57762]
[6 "chrIII" 261074]
[5 "chrIV" 340245]
[5 "chrIX" 723245]
[4 "chrV" 833233]
[5 "chrVI" 977468]
[6 "chrVII" 1045025]
[7 "chrVIII" 1317776]
[4 "chrX" 1458453]
[5 "chrXI" 1644907]
[6 "chrXII" 1811627]
[7 "chrXIII" 2081188]
[6 "chrXIV" 2312312]
[5 "chrXV" 2508412]
[6 "chrXVI" 2781251]
[4 "chrM" 3018284]]
The apparent consistency of these values give us some initial
confidence that we are reading the index correctly. Note, however, the
curious fact that chrIX appears between IV and V.
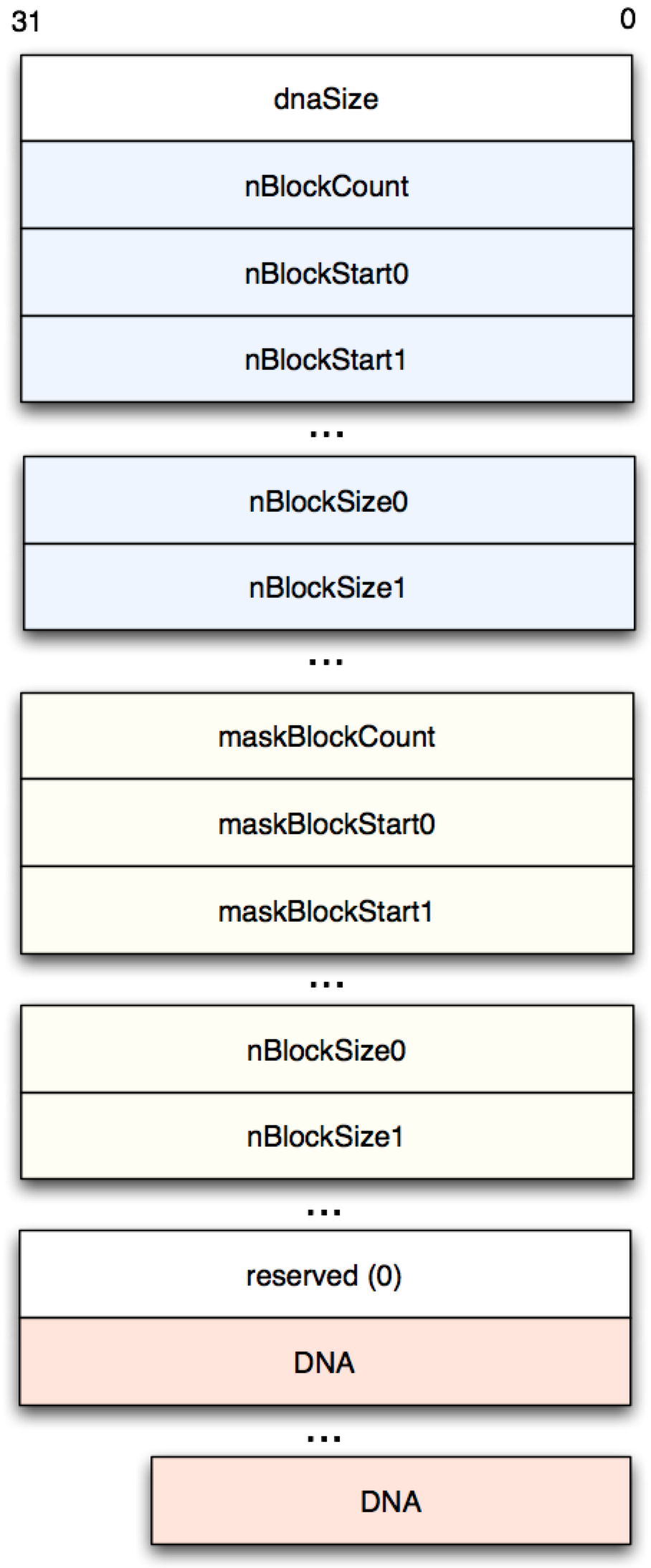
With this encouraging start, we can now attack the sequences proper. These are laid out as shown in Figure 2, with block metadata prepended to the actual DNA sequences:
The “N blocks” are blocks of unknown sequences with specified offsets
and lengths. Masked blocks are blocks which are known repetitions
(indicated as lower case “a”, “g”, “c” and “t” in the text-based
‘FASTA’ file format). We are obviously most interested in dnaSize, the
number of base pairs in the sequence, and the actual sequence values
themselves.
Unpacking the above data format (except the base pairs per se) makes
heavy use of get32, which just returns the unsigned 32-bit integer at
the specified file location. This code doesn’t need to be super
efficient, since the block headers themselves are quite small. The
metadata for the entire file is returned as a sequence of maps.
(defn getblk [fname offset n]
(let [ret (map #(get32 fname (+ offset (* 4 %))) (range n))
offset (skip offset n)]
[ret offset]))
(defn sequence-headers
"
Get sequence headers from .2bit file, as documented in
http://genome.ucsc.edu/FAQ/FAQformat#format7. Returns a list of maps
with details for each sequence.
"
[fname]
(let [seqcnt (file-header fname)]
(for [[nlen name ofs] (file-index fname seqcnt)]
(let [[[dna-size] ofs] (getblk fname ofs 1)
[[n-block-count] ofs] (getblk fname ofs 1)
[n-block-starts ofs] (getblk fname ofs n-block-count)
[n-block-sizes ofs] (getblk fname ofs n-block-count)
[[mask-block-count] ofs] (getblk fname ofs 1)
[mask-block-starts ofs] (getblk fname ofs mask-block-count)
[mask-block-sizes ofs] (getblk fname ofs mask-block-count)
[[reserved] ofs] (getblk fname ofs 1)]
(assert (zero? reserved))
{:name name
:nlen nlen
:dna-size dna-size
:n-block-count n-block-count
:n-block-starts n-block-starts
:n-block-sizes n-block-sizes
:mask-block-starts mask-block-starts
:mask-block-sizes mask-block-sizes
:dna-offset ofs}))))
A few sanity checks are included in test_core.clj to make sure we’re
decoding the metadata correctly. Requirement (3) is done!
The final step (Requirement (1)), is to actually get our base pairs
(BPs). Since we have to assume the data set is very large (as is the
case with the human genome), we cannot read the entire DNA sequence at
once. The first part is to break the dna-size base pairs (remember we
have 2 bits per BP, or 4 BP/byte), starting at dna-offset. First we
obtain the “coordinates” of the sequences we want to read:
(defn get-buffer-starts-and-lengths
"
Return buffer offsets (starting at ofs) required to cleanly read a
total of m bytes no more than n at a time
"
[ofs n m]
(loop [a ofs
len n
ret []]
(if (>= a (+ m ofs))
ret
(recur (+ a n)
n
(conj ret [a (min len (- (+ m ofs) a))])))))
;; Example:
(get-buffer-starts-and-lengths 100 200 512)
;=> [[100 200] [300 200] [500 112]]
At long last, having obtained the locations and lengths we want to read from, we can get our sequences out:
(defn genome-sequence
"
Read a specific sequence, or all sequences in a file concatenated
together; return it as a lazy seq.
"
([fname]
(let [sh (sequence-headers fname)]
(mapcat #(genome-sequence fname %1 %2)
(map :dna-offset sh)
(map :dna-size sh))))
([fname ofs dna-len]
(take dna-len
(apply concat
(let [byte-len (rounding-up-divide dna-len 4)
starts-and-lengths (get-buffer-starts-and-lengths
ofs 10000 byte-len)]
(for [[offset length] starts-and-lengths
:let [buf (read-with-offset fname offset length)]
b buf]
(byte-to-base-pairs b)))))))
;; Example:
(->> yeast
genome-sequence
(take 30))
;=> (:C :C :A :C :A :C :C :A :C :A :C :C :C :A :C
; :A :C :A :C :C :C :A :C :A :C :A :C :C :A :C)
This function allows us to choose the entire genome, or that for a given offset and number of base pairs (whether from the metadata for an entire chromosome, or some smaller region, thus satisfying (2)).
The next post will focus on verification of correctness of this code; subsequent posts will begin to explore the characteristics of this data for various genomes, human or otherwise.
Blog Posts (170)
Select from below, view all posts, or choose only posts for:art clojure code emacs lisp misc orgmode physics python ruby sketchup southpole writing
From Elegance to Speed code lisp clojure physics Wednesday, September 25, 2019
Common Lisp How-Tos code lisp Sunday, September 15, 2019
Implementing Scheme in Python code python lisp Friday, September 13, 2019
A Daily Journal in Org Mode writing code emacs Monday, September 2, 2019
Show at Northwestern University Prosthetics-Orthotics Center art Saturday, October 20, 2018
Color Notations art Thursday, September 20, 2018
Painting and Time art Tuesday, May 29, 2018
Learning Muscular Anatomy code clojure art emacs orgmode Thursday, February 22, 2018
Reflections on a Year of Daily Memory Drawings art Monday, September 18, 2017
Repainting art Sunday, May 14, 2017
Daily Memory Drawings art Tuesday, January 3, 2017
Questions to Ask art Thursday, February 25, 2016
Macro-writing Macros code clojure Wednesday, November 25, 2015
Time Limits art Saturday, April 25, 2015
Lazy Physics code clojure physics Thursday, February 12, 2015
Fun with Instaparse code clojure Tuesday, November 12, 2013
Nucleotide Repetition Lengths code clojure Sunday, November 3, 2013
Updating the Genome Decoder code clojure Saturday, July 13, 2013
Getting Our Hands Dirty (with the Human Genome) code clojure Wednesday, July 10, 2013
Validating the Genome Decoder code clojure Sunday, July 7, 2013
Exploratory Genomics with Clojure code clojure Friday, July 5, 2013
Rosalind Problems in Clojure code clojure Sunday, June 9, 2013
Introduction to Context Managers in Python code python Saturday, April 20, 2013
Processes vs. Threads for Integration Testing code python Friday, April 19, 2013
Resources for Learning Clojure code clojure Monday, May 21, 2012
Continuous Testing in Python, Clojure and Blub code clojure python Saturday, March 31, 2012
Programming Languages code clojure python Thursday, December 22, 2011
Milvans and Container Malls southpole Friday, December 2, 2011
Oxygen southpole Tuesday, November 29, 2011
Ghost southpole Monday, November 28, 2011
Turkey, Stuffing, Eclipse southpole Sunday, November 27, 2011
Wind Storm and Moon Dust southpole Friday, November 25, 2011
Shower Instructions southpole Wednesday, November 23, 2011
Fresh Air and Bananas southpole Sunday, November 20, 2011
Traveller and the Human Chain southpole Thursday, November 17, 2011
Reveille southpole Monday, November 14, 2011
Drifts southpole Sunday, November 13, 2011
Bon Voyage southpole Friday, November 11, 2011
A Nicer Guy? southpole Friday, November 11, 2011
The Quiet Earth southpole Monday, November 7, 2011
Ten southpole Tuesday, November 1, 2011
The Wheel art Wednesday, October 19, 2011
Plein Air art Saturday, August 27, 2011
ISO50 southpole art Thursday, July 28, 2011
SketchUp and MakeHuman sketchup art Monday, June 27, 2011
In Defense of Hobbies misc code art Sunday, May 29, 2011
Closure southpole Tuesday, January 25, 2011
Takeoff southpole Sunday, January 23, 2011
Mummification southpole Sunday, January 23, 2011
Eleventh Hour southpole Saturday, January 22, 2011
Diamond southpole Thursday, January 20, 2011
Baby, It's Cold Outside southpole Wednesday, January 19, 2011
Fruition southpole Tuesday, January 18, 2011
Friendly Radiation southpole Tuesday, January 18, 2011
A Place That Wants You Dead southpole Monday, January 17, 2011
Marathon southpole Sunday, January 16, 2011
Deep Fried Macaroni and Cheese Balls southpole Saturday, January 15, 2011
Retrograde southpole Thursday, January 13, 2011
Three southpole Wednesday, January 12, 2011
Transitions southpole Tuesday, January 11, 2011
The Future southpole Monday, January 10, 2011
Sunday southpole Sunday, January 9, 2011
Radio Waves southpole Saturday, January 8, 2011
Settling In southpole Thursday, January 6, 2011
Revolution Number Nine southpole Thursday, January 6, 2011
Red Eye to McMurdo southpole Wednesday, January 5, 2011
That's the Way southpole Tuesday, January 4, 2011
Faults in Ice and Rock southpole Monday, January 3, 2011
Bardo southpole Monday, January 3, 2011
Chasing the Sun southpole Sunday, January 2, 2011
Downhole southpole Monday, December 13, 2010
Coming Out of Hibernation southpole Monday, September 13, 2010
Managing the Most Remote Data Center in the World code southpole Friday, July 30, 2010
Ruby Plugins for Sketchup art sketchup ruby code Saturday, April 3, 2010
The Cruel Stranger misc Tuesday, November 10, 2009
Photoshop on a Dime art Monday, October 12, 2009
Man on Wire misc Friday, October 9, 2009
Videos southpole Friday, May 1, 2009
Posing Rigs art Sunday, April 5, 2009
Metric art Monday, March 2, 2009
Cuba southpole Sunday, February 22, 2009
Wickets southpole Monday, February 16, 2009
Safe southpole Sunday, February 15, 2009
Broken Glasses southpole Thursday, February 12, 2009
End of the Second Act southpole Friday, February 6, 2009
Pigs and Fish southpole Wednesday, February 4, 2009
Last Arrivals southpole Saturday, January 31, 2009
Lily White southpole Saturday, January 31, 2009
In a Dry and Waterless Place southpole Friday, January 30, 2009
Immortality southpole Thursday, January 29, 2009
Routine southpole Thursday, January 29, 2009
Tourists southpole Wednesday, January 28, 2009
Passing Notes southpole Thursday, January 22, 2009
Translation southpole Tuesday, January 20, 2009
RNZAF southpole Monday, January 19, 2009
The Usual Delays southpole Monday, January 19, 2009
CHC southpole Saturday, January 17, 2009
Wyeth on Another Planet art Saturday, January 17, 2009
Detox southpole Friday, January 16, 2009
Packing southpole Tuesday, January 13, 2009
Nails southpole Friday, January 9, 2009
Gearing Up southpole Tuesday, January 6, 2009
Gouache, and a new system for conquering the world art Sunday, November 30, 2008
Fall 2008 HPAC Studies art Friday, November 21, 2008
YABP (Yet Another Blog Platform) southpole Thursday, November 20, 2008
A Bath southpole Monday, February 18, 2008
Green Marathon southpole Saturday, February 16, 2008
Sprung southpole Friday, February 15, 2008
Outta Here southpole Wednesday, February 13, 2008
Lame Duck DAQer southpole Tuesday, February 12, 2008
Eclipse Town southpole Saturday, February 9, 2008
One More Week southpole Wednesday, February 6, 2008
IceCube Laboratory Video Tour; Midrats Finale southpole Friday, February 1, 2008
SPIFF, Party, Shift Change southpole Monday, January 28, 2008
Good things come in threes, or 18s southpole Saturday, January 26, 2008
Sleepless in the Station southpole Thursday, January 24, 2008
Post Deploy southpole Monday, January 21, 2008
IceCube and The Beatles southpole Saturday, January 19, 2008
Midrats southpole Saturday, January 19, 2008
Video: Flight to South Pole southpole Thursday, January 17, 2008
Almost There southpole Wednesday, January 16, 2008
The Pure Land southpole Wednesday, January 16, 2008
There are no mice in the Hotel California Bunkroom southpole Sunday, January 13, 2008
Short Timer southpole Sunday, January 13, 2008
Sleepy in MacTown southpole Saturday, January 12, 2008
Sir Ed southpole Friday, January 11, 2008
Pynchon, Redux southpole Friday, January 11, 2008
Superposition of Luggage States southpole Friday, January 11, 2008
Shortcut to Toast southpole Friday, January 11, 2008
Flights: Round 1 southpole Thursday, January 10, 2008
Packing for the Pole southpole Monday, January 7, 2008
Goals for Trip southpole Sunday, January 6, 2008
Balaklavas southpole Friday, January 4, 2008
Tree and Man (Test Post) southpole Friday, December 28, 2007
Schedule southpole Sunday, December 16, 2007
How to mail stuff to John at the South Pole southpole Sunday, November 25, 2007
Summer and Winter southpole Tuesday, March 6, 2007
Homeward Bound southpole Thursday, February 22, 2007
Redeployment southpole Monday, February 19, 2007
Short-timer southpole Sunday, February 18, 2007
The Cleanest Air in the World southpole Saturday, February 17, 2007
One more day (?) southpole Friday, February 16, 2007
One more week (?) southpole Thursday, February 15, 2007
Closing Softly southpole Monday, February 12, 2007
More Photos southpole Friday, February 9, 2007
Super Bowl Wednesday southpole Thursday, February 8, 2007
Night Owls southpole Tuesday, February 6, 2007
First Week southpole Friday, February 2, 2007
More Ice Pix southpole Wednesday, January 31, 2007
Settling In southpole Tuesday, January 30, 2007
NPX southpole Monday, January 29, 2007
Pole Bound southpole Sunday, January 28, 2007
Bad Dirt southpole Saturday, January 27, 2007
The Last Laugh southpole Friday, January 26, 2007
First Delay southpole Thursday, January 25, 2007
Nope southpole Thursday, January 25, 2007
Batteries and Sheep southpole Wednesday, January 24, 2007
All for McNaught southpole Tuesday, January 23, 2007
t=0 southpole Monday, January 22, 2007
The Big (Really really big...) Picture southpole Monday, January 22, 2007
Giacometti southpole Monday, January 22, 2007
Descent southpole Monday, January 22, 2007
Video Tour southpole Saturday, January 20, 2007
How to subscribe to blog updates southpole Monday, December 11, 2006
What The Blog is For southpole Sunday, December 10, 2006
Auckland southpole Tuesday, January 11, 2005
Halfway Around the World; Dragging the Soul Behind southpole Monday, January 10, 2005
Launched southpole Sunday, January 9, 2005
Getting Ready (t minus 2 days) southpole Friday, January 7, 2005
Subscribe: RSS feed ... all topics ... or Clojure only / Lisp only