Getting Our Hands Dirty (with the Human Genome)
code clojure
Wednesday, July 10, 2013
In the past two posts, we created and validated a decoder for genome data in 2bit format.
Let’s start actually looking at the human and yeast genomes per se. First, if you haven’t downloaded it yet, you’ll need a copy of the human genome data file:
cd /tmp wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/bigZips/hg19.2bit
(Depending on your local bandwidth conditions, this can take quite awhile.)
Again we define convenience vars for the two data files:
(def human "/tmp/hg19.2bit") (def yeast (as-file (resource "sacCer3.2bit")))
The simplest thing we can do is count base pairs:
jenome.core> (time (->> yeast genome-sequence count)) "Elapsed time: 17577.042 msecs" 12157105 jenome.core> (time (->> human genome-sequence count)) "Elapsed time: 4513537.422 msecs" -1157806032
WTF?! count has overflowed (if you convert -1157806032 to an unsigned int you get 3137161264, which makes more sense). This, and our running time of 75 minutes, is our first hint of pain associated with handling with such a large dataset.
It’s worth noting that number overflows in Clojure generally raise an ArithmeticException (or silently get promoted to BigIntegers, depending on the operators used) – I emailed the Clojure mailing list about this surprise and a ticket has been made.
The overflow is easily enough remedied with a non-overflowing count
(=inc’= is one of the aforementioned promoting operators):
(defn count' [s] ;; Regular count overflows to negative int!
(loop [s s, n 0]
(if (seq s)
(recur (rest s)
(inc' n))
n)))
Which gives us the unsigned version of our previous answer, in 10% more time:
jenome.core> (time (->> human genome-sequence count')) "Elapsed time: 4931630.301 msecs" 3137161264 jenome.core> (float (/ 3137161264 12157105)) 258.05167
(Update 2/14/2014: Matthew Wampler-Doty came up with a more elegant =count’=:
(defn count' [s] (reduce (fn [x _] (inc x)) 0 s))
which is about 25% faster than my slightly more naïve version.)
We now have our first bit of insight into human vs. yeast genomes: we humans have about 260x more base pairs than yeast do.
The next question is, What are the relative proportions of occurrence
of each base pair? Easily answered by the frequencies function:
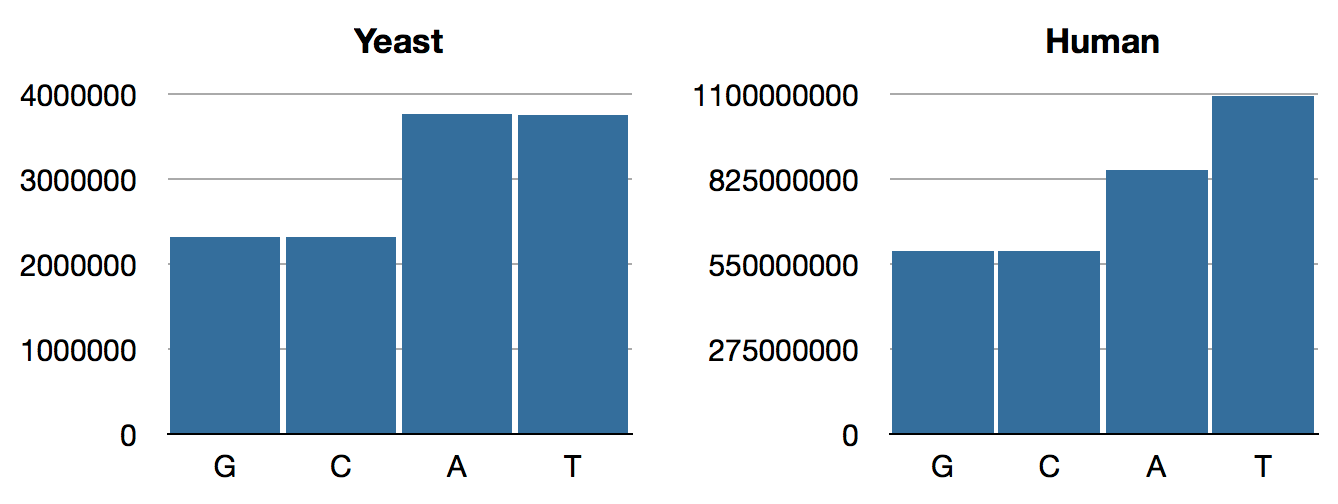
jenome.core> (->> yeast genome-sequence frequencies)
{:C 2320576, :A 3766349, :T 3753080, :G 2317100}
jenome.core> (->> human genome-sequence frequencies)
{:T 1095906163, :A 854963149, :C 592966724, :G 593325228}
jenome.core>
Those numbers have a lot of digits, so here are some bar charts:
Though we are playing ignorant data analysts rather than real biologists, this looks a bit fishy right out of the gate. Why so many Ts?
As a sanity check, perhaps we ought to look more closely at the actual data:
jenome.core> (->>
human
genome-sequence
(map name)
(take 100)
(apply str))
;=> "TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT
; TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT"
That doesn’t look good at all. How are the first 10000 base pairs distributed?
jenome.core> (->>
human
genome-sequence
(take 10000)
frequencies)
;=> {:T 10000}
How about the next 10,0000?
jenome.core> (->>
human
genome-sequence
(drop 10000)
(take 10000)
frequencies)
;=> {:T 2012, :A 2065, :C 3028, :G 2895}
That looks a lot better. Playing around with this shows that exactly the first 10,000 base pairs are Ts.
The astute reader will recall that the file index in our decoder had slots for unknown (“N-block”) sequences. Though we didn’t explore them explicitly, /S. Cerviciae/’s genome had nothing in these slots. It’s time we went back and looked at our human’s file index:
(->> human
sequence-headers
(map (juxt :name :dna-size :n-block-count))
clojure.pprint/pprint)
;; =>
(["chr1" 249250621 39]
["chr2" 243199373 24]
["chr3" 198022430 9]
["chr4" 191154276 12]
["chr5" 180915260 7]
["chr6" 171115067 11]
["chr7" 159138663 17]
["chrX" 155270560 23]
["chr8" 146364022 9]
;; LOTS more entries...
)
Holy cow, they all have N-blocks. This is only the first section; this Gist shows the entire set of N-blocks for all the sequences. (It also shows that there are many more files in the index than the yeast genome had, and many more than one for each of the 24 human chromosomes – clearly there are many things to learn about this data.)
Moreover, if we look at the N-blocks for the first sequence:
(->> human
sequence-headers
first
((juxt :n-block-starts :n-block-sizes))
(apply interleave)
(partition 2)
(map vec)
clojure.pprint/pprint)
;; Gives:
([0 10000]
[177417 50000]
[267719 50000]
[471368 50000]
;; ... total of 39 entries.
)
As expected, our first 10,000 base pairs are actually unknown. The 2-bit file format apparently uses 0’s for the unknowns AND to represent Ts. The rest of the offsets and lengths are in this Gist. (Perhaps someone can explain to me why these are all multiples of 10000!)
All this means that our decoder needs a tweak to give us :N instead
of :T when the position of the base pair is inside one of the
N-blocks. This we will accomplish in the next post, before we proceed
to the study of other properties of the genome.
Before we end, it’s worth noting that, had we been real biologists, we would surely have known about Chargaff’s Rule, which states that A and T ratios should be the same, as are G and C, as born out in our yeast distribution, above.
Though we haven’t gained mastery over too much of the genome yet, it
is worth pointing out Clojure’s strengths in the above and previous
posts. With comparatively little code we are able to investigate many
of the features of these data sets, thanks to the power of the
threading macro ->>, higher-order functions such as juxt and map, and
sequence handling functions such as interleave, partition, and
frequencies. We’ve also processed large data sets “lazily” without
needing to fit large amounts of data in memory at once. And we have
hardly scratched the surface of Clojure’s tools for parallel
computation, though we may make more use of these in coming posts.
Next: Updating the Decoder
Blog Posts (170)
Select from below, view all posts, or choose only posts for:art clojure code emacs lisp misc orgmode physics python ruby sketchup southpole writing
From Elegance to Speed code lisp clojure physics Wednesday, September 25, 2019
Common Lisp How-Tos code lisp Sunday, September 15, 2019
Implementing Scheme in Python code python lisp Friday, September 13, 2019
A Daily Journal in Org Mode writing code emacs Monday, September 2, 2019
Show at Northwestern University Prosthetics-Orthotics Center art Saturday, October 20, 2018
Color Notations art Thursday, September 20, 2018
Painting and Time art Tuesday, May 29, 2018
Learning Muscular Anatomy code clojure art emacs orgmode Thursday, February 22, 2018
Reflections on a Year of Daily Memory Drawings art Monday, September 18, 2017
Repainting art Sunday, May 14, 2017
Daily Memory Drawings art Tuesday, January 3, 2017
Questions to Ask art Thursday, February 25, 2016
Macro-writing Macros code clojure Wednesday, November 25, 2015
Time Limits art Saturday, April 25, 2015
Lazy Physics code clojure physics Thursday, February 12, 2015
Fun with Instaparse code clojure Tuesday, November 12, 2013
Nucleotide Repetition Lengths code clojure Sunday, November 3, 2013
Updating the Genome Decoder code clojure Saturday, July 13, 2013
Validating the Genome Decoder code clojure Sunday, July 7, 2013
A Two Bit Decoder code clojure Saturday, July 6, 2013
Exploratory Genomics with Clojure code clojure Friday, July 5, 2013
Rosalind Problems in Clojure code clojure Sunday, June 9, 2013
Introduction to Context Managers in Python code python Saturday, April 20, 2013
Processes vs. Threads for Integration Testing code python Friday, April 19, 2013
Resources for Learning Clojure code clojure Monday, May 21, 2012
Continuous Testing in Python, Clojure and Blub code clojure python Saturday, March 31, 2012
Programming Languages code clojure python Thursday, December 22, 2011
Milvans and Container Malls southpole Friday, December 2, 2011
Oxygen southpole Tuesday, November 29, 2011
Ghost southpole Monday, November 28, 2011
Turkey, Stuffing, Eclipse southpole Sunday, November 27, 2011
Wind Storm and Moon Dust southpole Friday, November 25, 2011
Shower Instructions southpole Wednesday, November 23, 2011
Fresh Air and Bananas southpole Sunday, November 20, 2011
Traveller and the Human Chain southpole Thursday, November 17, 2011
Reveille southpole Monday, November 14, 2011
Drifts southpole Sunday, November 13, 2011
Bon Voyage southpole Friday, November 11, 2011
A Nicer Guy? southpole Friday, November 11, 2011
The Quiet Earth southpole Monday, November 7, 2011
Ten southpole Tuesday, November 1, 2011
The Wheel art Wednesday, October 19, 2011
Plein Air art Saturday, August 27, 2011
ISO50 southpole art Thursday, July 28, 2011
SketchUp and MakeHuman sketchup art Monday, June 27, 2011
In Defense of Hobbies misc code art Sunday, May 29, 2011
Closure southpole Tuesday, January 25, 2011
Takeoff southpole Sunday, January 23, 2011
Mummification southpole Sunday, January 23, 2011
Eleventh Hour southpole Saturday, January 22, 2011
Diamond southpole Thursday, January 20, 2011
Baby, It's Cold Outside southpole Wednesday, January 19, 2011
Fruition southpole Tuesday, January 18, 2011
Friendly Radiation southpole Tuesday, January 18, 2011
A Place That Wants You Dead southpole Monday, January 17, 2011
Marathon southpole Sunday, January 16, 2011
Deep Fried Macaroni and Cheese Balls southpole Saturday, January 15, 2011
Retrograde southpole Thursday, January 13, 2011
Three southpole Wednesday, January 12, 2011
Transitions southpole Tuesday, January 11, 2011
The Future southpole Monday, January 10, 2011
Sunday southpole Sunday, January 9, 2011
Radio Waves southpole Saturday, January 8, 2011
Settling In southpole Thursday, January 6, 2011
Revolution Number Nine southpole Thursday, January 6, 2011
Red Eye to McMurdo southpole Wednesday, January 5, 2011
That's the Way southpole Tuesday, January 4, 2011
Faults in Ice and Rock southpole Monday, January 3, 2011
Bardo southpole Monday, January 3, 2011
Chasing the Sun southpole Sunday, January 2, 2011
Downhole southpole Monday, December 13, 2010
Coming Out of Hibernation southpole Monday, September 13, 2010
Managing the Most Remote Data Center in the World code southpole Friday, July 30, 2010
Ruby Plugins for Sketchup art sketchup ruby code Saturday, April 3, 2010
The Cruel Stranger misc Tuesday, November 10, 2009
Photoshop on a Dime art Monday, October 12, 2009
Man on Wire misc Friday, October 9, 2009
Videos southpole Friday, May 1, 2009
Posing Rigs art Sunday, April 5, 2009
Metric art Monday, March 2, 2009
Cuba southpole Sunday, February 22, 2009
Wickets southpole Monday, February 16, 2009
Safe southpole Sunday, February 15, 2009
Broken Glasses southpole Thursday, February 12, 2009
End of the Second Act southpole Friday, February 6, 2009
Pigs and Fish southpole Wednesday, February 4, 2009
Last Arrivals southpole Saturday, January 31, 2009
Lily White southpole Saturday, January 31, 2009
In a Dry and Waterless Place southpole Friday, January 30, 2009
Immortality southpole Thursday, January 29, 2009
Routine southpole Thursday, January 29, 2009
Tourists southpole Wednesday, January 28, 2009
Passing Notes southpole Thursday, January 22, 2009
Translation southpole Tuesday, January 20, 2009
RNZAF southpole Monday, January 19, 2009
The Usual Delays southpole Monday, January 19, 2009
CHC southpole Saturday, January 17, 2009
Wyeth on Another Planet art Saturday, January 17, 2009
Detox southpole Friday, January 16, 2009
Packing southpole Tuesday, January 13, 2009
Nails southpole Friday, January 9, 2009
Gearing Up southpole Tuesday, January 6, 2009
Gouache, and a new system for conquering the world art Sunday, November 30, 2008
Fall 2008 HPAC Studies art Friday, November 21, 2008
YABP (Yet Another Blog Platform) southpole Thursday, November 20, 2008
A Bath southpole Monday, February 18, 2008
Green Marathon southpole Saturday, February 16, 2008
Sprung southpole Friday, February 15, 2008
Outta Here southpole Wednesday, February 13, 2008
Lame Duck DAQer southpole Tuesday, February 12, 2008
Eclipse Town southpole Saturday, February 9, 2008
One More Week southpole Wednesday, February 6, 2008
IceCube Laboratory Video Tour; Midrats Finale southpole Friday, February 1, 2008
SPIFF, Party, Shift Change southpole Monday, January 28, 2008
Good things come in threes, or 18s southpole Saturday, January 26, 2008
Sleepless in the Station southpole Thursday, January 24, 2008
Post Deploy southpole Monday, January 21, 2008
IceCube and The Beatles southpole Saturday, January 19, 2008
Midrats southpole Saturday, January 19, 2008
Video: Flight to South Pole southpole Thursday, January 17, 2008
Almost There southpole Wednesday, January 16, 2008
The Pure Land southpole Wednesday, January 16, 2008
There are no mice in the Hotel California Bunkroom southpole Sunday, January 13, 2008
Short Timer southpole Sunday, January 13, 2008
Sleepy in MacTown southpole Saturday, January 12, 2008
Sir Ed southpole Friday, January 11, 2008
Pynchon, Redux southpole Friday, January 11, 2008
Superposition of Luggage States southpole Friday, January 11, 2008
Shortcut to Toast southpole Friday, January 11, 2008
Flights: Round 1 southpole Thursday, January 10, 2008
Packing for the Pole southpole Monday, January 7, 2008
Goals for Trip southpole Sunday, January 6, 2008
Balaklavas southpole Friday, January 4, 2008
Tree and Man (Test Post) southpole Friday, December 28, 2007
Schedule southpole Sunday, December 16, 2007
How to mail stuff to John at the South Pole southpole Sunday, November 25, 2007
Summer and Winter southpole Tuesday, March 6, 2007
Homeward Bound southpole Thursday, February 22, 2007
Redeployment southpole Monday, February 19, 2007
Short-timer southpole Sunday, February 18, 2007
The Cleanest Air in the World southpole Saturday, February 17, 2007
One more day (?) southpole Friday, February 16, 2007
One more week (?) southpole Thursday, February 15, 2007
Closing Softly southpole Monday, February 12, 2007
More Photos southpole Friday, February 9, 2007
Super Bowl Wednesday southpole Thursday, February 8, 2007
Night Owls southpole Tuesday, February 6, 2007
First Week southpole Friday, February 2, 2007
More Ice Pix southpole Wednesday, January 31, 2007
Settling In southpole Tuesday, January 30, 2007
NPX southpole Monday, January 29, 2007
Pole Bound southpole Sunday, January 28, 2007
Bad Dirt southpole Saturday, January 27, 2007
The Last Laugh southpole Friday, January 26, 2007
First Delay southpole Thursday, January 25, 2007
Nope southpole Thursday, January 25, 2007
Batteries and Sheep southpole Wednesday, January 24, 2007
All for McNaught southpole Tuesday, January 23, 2007
t=0 southpole Monday, January 22, 2007
The Big (Really really big...) Picture southpole Monday, January 22, 2007
Giacometti southpole Monday, January 22, 2007
Descent southpole Monday, January 22, 2007
Video Tour southpole Saturday, January 20, 2007
How to subscribe to blog updates southpole Monday, December 11, 2006
What The Blog is For southpole Sunday, December 10, 2006
Auckland southpole Tuesday, January 11, 2005
Halfway Around the World; Dragging the Soul Behind southpole Monday, January 10, 2005
Launched southpole Sunday, January 9, 2005
Getting Ready (t minus 2 days) southpole Friday, January 7, 2005
Subscribe: RSS feed ... all topics ... or Clojure only / Lisp only